Aldolase A
| ALDOA | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | ALDOA, ALDA, GSD12, HEL-S-87p, aldolase, fructose-bisphosphate A, Aldolase A | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 103850; MGI: 87994; HomoloGene: 141054; GeneCards: ALDOA; OMA:ALDOA - orthologs | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| fructose-bisphosphate aldolase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC no. | 4.1.2.13 | ||||||||
| CAS no. | 9024-52-6 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
Aldolase A (ALDOA, or ALDA), also known as fructose-bisphosphate aldolase, is an enzyme that in humans is encoded by the ALDOA gene on chromosome 16.
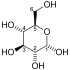
The protein encoded by this gene is a glycolytic enzyme that catalyzes the reversible conversion of fructose-1,6-bisphosphate to glyceraldehyde 3-phosphate (G3P) and dihydroxyacetone phosphate (DHAP). Three aldolase isozymes (A, B, and C), encoded by three different genes, are differentially expressed during development. Aldolase A is found in the developing embryo and is produced in even greater amounts in adult muscle. Aldolase A expression is repressed in adult liver, kidney and intestine and similar to aldolase C levels in brain and other nervous tissue. Aldolase A deficiency has been associated with myopathy and hemolytic anemia. Alternative splicing and alternative promoter usage results in multiple transcript variants. Related pseudogenes have been identified on chromosomes 3 and 10.[5]
Structure
[edit]ALDOA is a homotetramer and one of the three aldolase isozymes (A, B, and C), encoded by three different genes.[6][7] The ALDOA gene contains 8 exons and the 5' UTR IB.[7] Key amino acids responsible for its catalytic function have been identified. The residue Tyr363 functions as the acid–base catalyst for protonating C3 of the substrate, while Lys146 is proposed to stabilize the negative charge of the resulting conjugate base of Tyr363 and the strained configuration of the C-terminal. Residue Glu187 participates in multiple functions, including FBP aldolase catalysis, acid–base catalysis during substrate binding, dehydration, and substrate cleavage.[8] Though ALDOA localizes to the nucleus, it lacks any known nuclear localization signals (NLS).[9]
Mechanism
[edit]In mammalian aldolase, the key catalytic amino acid residues involved in the reaction are lysine and tyrosine. The tyrosine acts as an efficient hydrogen acceptor while the lysine covalently binds and stabilizes the intermediates. Many bacteria use two magnesium ions in place of the lysine. [citation needed]
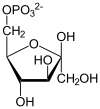
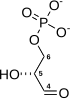
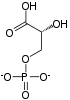
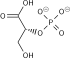
| β-D-fructose 1,6-phosphate | fructose-bisphosphate aldolase | D-glyceraldehyde 3-phosphate | dihydroxyacetone phosphate | ||

|

|
+ | 
| ||

| |||||
Compound C05378 at KEGG Pathway Database. Enzyme 4.1.2.13 at KEGG Pathway Database. Compound C00111 at KEGG Pathway Database. Compound C00118 at KEGG Pathway Database.
The numbering of the carbon atoms indicates the fate of the carbons according to their position in fructose 6-phosphate.
Function
[edit]ALDOA is a key enzyme in the fourth step of glycolysis, as well as in the reverse pathway gluconeogenesis. It catalyzes the reversible conversion of fructose-1,6-bisphosphate to glyceraldehydes-3-phosphate and dihydroxyacetone phosphate by aldol cleavage of the C3–C4 bond. As a result, it is a crucial player in ATP biosynthesis.[6][8][9][10][11] ALDOA also contributes to other "moonlighting" functions such as muscle maintenance, regulation of cell shape and motility, striated muscle contraction, actin cytoskeleton organization, and regulation of cell proliferation.[6][9][10] ALDOA likely regulates actin cytoskeleton remodeling through interacting with cytohesin-2 (ARNO) and Arf6.[10]
ALDOA is ubiquitously expressed in most tissues, though it is predominantly expressed in developing embryo and adult muscle.[6][11] In lymphocytes, ALDOA is the predominant aldolase isoform.[11] Within the cell, ALDOA typically localizes to the cytoplasm, but it can localize to the nucleus during DNA synthesis of the cell cycle S phase. This nuclear localization is regulated by the protein kinases AKT and p38. It is suggested that the nucleus serves as a reservoir for ALDOA in low glucose conditions.[9] ALDOA has also been found in mitochondria.[11]
ALDOA is regulated by the energy metabolism substrates glucose, lactate, and glutamine.[9] In human mast cells (MCs), ALDOA has been observed to undergo post-translational regulation by protein tyrosine nitration, which may alter its relative affinity for FBP and/or IP3. This change then affects IP3 and PLC signaling cascades in IgE-dependent responses.[11]
Clinical significance
[edit]Aldolase A (ALDOA) is highly expressed in multiple cancers, including lung squamous cell carcinoma (LSCC), renal cancer, and hepatocellular carcinoma. It is proposed that ALDOA overexpression enhances glycolysis in these tumor cells, promoting their growth. In LSCC, its upregulation correlates with metastasis and poor prognosis, while its downregulation reduces tumor cell motility and tumorigenesis. Thus, ALDOA could be a potential LSCC biomarker and therapeutic drug target.[6]
Aldolase A deficiency is a rare, autosomal recessive disorder that is linked to hemolysis and accompanied by weakness, muscle pain, and myopathy.[7]
Interactive pathway map
[edit]Click on genes, proteins and metabolites below to link to respective articles.[§ 1]
- ^ The interactive pathway map can be edited at WikiPathways: "GlycolysisGluconeogenesis_WP534".
Interactions
[edit]Aldolase A has been shown to interact with:
- PLD2,[12]
- actin,[11]
- GLUT4,[13]
- phospholipase D2,[13]
- light chain 8 of dynein,[13]
- erythrocyte anion exchanger Band 3 protein,[13]
- ryanodine receptor,[11]
- Cytohesin-2,[10] and
- V-ATPase (vacuolar-type H+-ATPase).[10]
See also
[edit]References
[edit]- ^ a b c GRCh38: Ensembl release 89: ENSG00000149925 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000030695 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Entrez Gene: ALDOA aldolase A, fructose-bisphosphate".
- ^ a b c d e Du S, Guan Z, Hao L, Song Y, Wang L, Gong L, Liu L, Qi X, Hou Z, Shao S (2014). "Fructose-bisphosphate aldolase a is a potential metastasis-associated marker of lung squamous cell carcinoma and promotes lung cell tumorigenesis and migration". PLOS ONE. 9 (1): e85804. Bibcode:2014PLoSO...985804D. doi:10.1371/journal.pone.0085804. PMC 3900443. PMID 24465716.
- ^ a b c Yao DC, Tolan DR, Murray MF, Harris DJ, Darras BT, Geva A, Neufeld EJ (15 March 2004). "Hemolytic anemia and severe rhabdomyolysis caused by compound heterozygous mutations of the gene for erythrocyte/muscle isozyme of aldolase, ALDOA(Arg303X/Cys338Tyr)". Blood. 103 (6): 2401–3. doi:10.1182/blood-2003-09-3160. PMID 14615364.
- ^ a b Tittmann K (December 2014). "Sweet siblings with different faces: the mechanisms of FBP and F6P aldolase, transaldolase, transketolase and phosphoketolase revisited in light of recent structural data". Bioorganic Chemistry. 57: 263–80. doi:10.1016/j.bioorg.2014.09.001. PMID 25267444.
- ^ a b c d e Mamczur P, Gamian A, Kolodziej J, Dziegiel P, Rakus D (December 2013). "Nuclear localization of aldolase A correlates with cell proliferation". Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 1833 (12): 2812–22. doi:10.1016/j.bbamcr.2013.07.013. PMID 23886627.
- ^ a b c d e Merkulova M, Hurtado-Lorenzo A, Hosokawa H, Zhuang Z, Brown D, Ausiello DA, Marshansky V (June 2011). "Aldolase directly interacts with ARNO and modulates cell morphology and acidic vesicle distribution". American Journal of Physiology. Cell Physiology. 300 (6): C1442-55. doi:10.1152/ajpcell.00076.2010. PMC 3118619. PMID 21307348.
- ^ a b c d e f g Sekar Y, Moon TC, Slupsky CM, Befus AD (1 July 2010). "Protein tyrosine nitration of aldolase in mast cells: a plausible pathway in nitric oxide-mediated regulation of mast cell function". Journal of Immunology. 185 (1): 578–87. doi:10.4049/jimmunol.0902720. PMID 20511553.
- ^ Kim JH, Lee Sukmook, Kim Jung Hwan, Lee Taehoon G, Hirata Masato, Suh Pann-Ghill, Ryu Sung Ho (Mar 2002). "Phospholipase D2 directly interacts with aldolase via Its PH domain". Biochemistry. 41 (10). United States: 3414–21. doi:10.1021/bi015700a. ISSN 0006-2960. PMID 11876650.
- ^ a b c d St-Jean M, Izard T, Sygusch J (11 May 2007). "A hydrophobic pocket in the active site of glycolytic aldolase mediates interactions with Wiskott-Aldrich syndrome protein". The Journal of Biological Chemistry. 282 (19): 14309–15. doi:10.1074/jbc.m611505200. PMID 17329259.
Further reading
[edit]- Pfleiderer G, Thöner M, Wachsmuth ED (1976). "Histological examination of the aldolase monomer composition of cells from human kidney and hypernephroid carcinoma". Beiträge zur Pathologie. 156 (3): 266–79. doi:10.1016/s0005-8165(75)80166-1. PMID 766744.
- Rehbein-Thöner M, Pfleiderer G (1977). "The changes in aldolase isoenzyme pattern during development of the human kidney and small intestine--demonstrated in organ extracts and tissue sections". Hoppe-Seyler's Z. Physiol. Chem. 358 (2): 169–80. doi:10.1515/bchm2.1977.358.1.169. PMID 844801.
- Wachsmuth ED (1976). "Differentiation of epithelial cells in human jejunum: localization and quantification of aminopeptidase, alkaline phosphatase and aldolase isozymes in tissue sections". Histochemistry. 48 (2): 101–9. doi:10.1007/BF00494548. PMID 955981. S2CID 6347675.
- Lee KN, Maxwell MD, Patterson MK, et al. (1992). "Identification of transglutaminase substrates in HT29 colon cancer cells: use of 5-(biotinamido)pentylamine as a transglutaminase-specific probe". Biochim. Biophys. Acta. 1136 (1): 12–6. doi:10.1016/0167-4889(92)90078-P. PMID 1353685.
- Dawson SJ, White LA (1992). "Treatment of Haemophilus aphrophilus endocarditis with ciprofloxacin". J. Infect. 24 (3): 317–20. doi:10.1016/S0163-4453(05)80037-4. PMID 1602151.
- Mukai T, Arai Y, Yatsuki H, et al. (1991). "An additional promoter functions in the human aldolase A gene, but not in rat". Eur. J. Biochem. 195 (3): 781–7. doi:10.1111/j.1432-1033.1991.tb15766.x. PMID 1999195.
- Gamblin SJ, Davies GJ, Grimes JM, et al. (1991). "Activity and specificity of human aldolases". J. Mol. Biol. 219 (4): 573–6. doi:10.1016/0022-2836(91)90650-U. PMID 2056525.
- Vértessy BG, Orosz F, Ovádi J (1991). "Modulation of the interaction between aldolase and glycerol-phosphate dehydrogenase by fructose phosphates". Biochim. Biophys. Acta. 1078 (2): 236–42. doi:10.1016/0167-4838(91)90564-g. PMID 2065091.
- Takasaki Y, Takahashi I, Mukai T, Hori K (1990). "Human aldolase A of a hemolytic anemia patient with Asp-128----Gly substitution: characteristics of an enzyme generated in E. coli transfected with the expression plasmid pHAAD128G". J. Biochem. 108 (2): 153–7. doi:10.1093/oxfordjournals.jbchem.a123174. PMID 2229018.
- Gamblin SJ, Cooper B, Millar JR, et al. (1990). "The crystal structure of human muscle aldolase at 3.0 A resolution". FEBS Lett. 262 (2): 282–6. doi:10.1016/0014-5793(90)80211-Z. PMID 2335208. S2CID 46133456.
- Kishi H, Mukai T, Hirono A, et al. (1988). "Human aldolase A deficiency associated with a hemolytic anemia: thermolabile aldolase due to a single base mutation". Proc. Natl. Acad. Sci. U.S.A. 84 (23): 8623–7. Bibcode:1987PNAS...84.8623K. doi:10.1073/pnas.84.23.8623. PMC 299598. PMID 2825199.
- Izzo P, Costanzo P, Lupo A, et al. (1987). "A new human species of aldolase A mRNA from fibroblasts". Eur. J. Biochem. 164 (1): 9–13. doi:10.1111/j.1432-1033.1987.tb10984.x. PMID 3030757.
- Inagaki H, Haimoto H, Hosoda S, Kato K (1988). "Aldolase C is localized in neuroendocrine cells". Experientia. 44 (9): 749–51. doi:10.1007/BF01959149. PMID 3046960. S2CID 10109239.
- Freemont PS, Dunbar B, Fothergill-Gilmore LA (1988). "The complete amino acid sequence of human skeletal-muscle fructose-bisphosphate aldolase". Biochem. J. 249 (3): 779–88. doi:10.1042/bj2490779. PMC 1148774. PMID 3355497.
- Izzo P, Costanzo P, Lupo A, et al. (1988). "Human aldolase A gene. Structural organization and tissue-specific expression by multiple promoters and alternate mRNA processing". Eur. J. Biochem. 174 (4): 569–78. doi:10.1111/j.1432-1033.1988.tb14136.x. PMID 3391172.
- Maire P, Gautron S, Hakim V, et al. (1988). "Characterization of three optional promoters in the 5' region of the human aldolase A gene". J. Mol. Biol. 197 (3): 425–38. doi:10.1016/0022-2836(87)90556-0. PMID 3441006.
- Kukita A, Yoshida MC, Fukushige S, et al. (1987). "Molecular gene mapping of human aldolase A (ALDOA) gene to chromosome 16". Hum. Genet. 76 (1): 20–6. doi:10.1007/BF00283044. PMID 3570299. S2CID 162055.
- Tolan DR, Niclas J, Bruce BD, Lebo RV (1987). "Evolutionary implications of the human aldolase-A, -B, -C, and -pseudogene chromosome locations". Am. J. Hum. Genet. 41 (5): 907–24. PMC 1684339. PMID 3674018.
- Sakakibara M, Mukai T, Hori K (1985). "Nucleotide sequence of a cDNA clone for human aldolase: a messenger RNA in the liver". Biochem. Biophys. Res. Commun. 131 (1): 413–20. doi:10.1016/0006-291X(85)91818-2. PMID 3840020.
- Ovádi J, Mohamed Osman IR, Batke J (1983). "Interaction of the dissociable glycerol-3-phosphate dehydrogenase and fructose-1,6-bisphosphate aldolase. Quantitative analysis by an extrinsic fluorescence probe". Eur. J. Biochem. 133 (2): 433–7. doi:10.1111/j.1432-1033.1983.tb07482.x. PMID 6406231.
External links
[edit]- http://pdbdev.sdsc.edu:48346/pdb/molecules/pdb50_5.html[permanent dead link]
- Fructose-Bisphosphate+Aldolase at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- ALDOA
- EC 4.1.2.13
- Human ALDOA genome location and ALDOA gene details page in the UCSC Genome Browser.
- PDBe-KB provides an overview of all the structure information available in the PDB for Human Fructose-bisphosphate aldolase A